Publications
WastewaterSCAN’s published body of work
An overview of the program’s peer-reviewed scientific literature, organized by topics and themes
WastewaterSCAN is a national leader in advancing the science of wastewater-based epidemiology (WBE) to generate actionable public-health insights. Since the program’s inception during the early days of the COVID-19 pandemic in 2020, WastewaterSCAN has led WBE’s evolution from an obscure, occasionally used scientific tool to a rigorous, widely accepted approach for comprehensive disease monitoring. WastewaterSCAN has published all of its findings in peer-reviewed scientific journals, many with collaborators from other academic institutions located worldwide, employees of CDC, and experts from local and state public health departments and wastewater utilities, underscoring the program’s commitment to facilitating adoption and routine use of WBE tools by public health agencies around the world.
To date, WastewaterSCAN has published more than 30 peer-reviewed manuscripts – virtually all of them accessible in free, open-source formats. One of the program’s oldest papers – a 2020 manuscript that compared SARS-CoV-2 concentrations recovered from wastewater liquids vs. primary settled solids – has been cited more than 300 times. WastewaterSCAN’s published body of work can be divided into two main categories: 1) Studies supporting the development of wastewater-based disease monitoring methods, and 2) studies that use the wastewater methods to generate public health insights. This resource, which serves as a comprehensive repository of WastewaterSCAN’s published body of work, is organized around these two main categories. The two main categories are further subdivided to highlight additional themes and attributes that various WastewaterSCAN papers have in common.

1. Developing wastewater-based methods for monitoring disease
At the beginning of the COVID-19 pandemic, WBE was an obscure, underdeveloped disease monitoring tool. Today, it’s a robust, widely vetted tool for monitoring more than a dozen disease targets, and WastewaterSCAN has played a leading role in making these advances possible. Shortly after the pandemic’s onset in 2019, the researchers behind WastewaterSCAN recognized that scientists would need to provide the public health community with reliable, standardized wastewater monitoring methods – methods that would pave the way to apply WBE science to monitor a range of disease targets. WastewaterSCAN soon emerged at the international forefront of wastewater method development, designing and vetting a suite of wastewater collection, processing and analysis protocols. Along the way, WastewaterSCAN demonstrated the advantages of monitoring wastewater solids instead of liquids – a method that has been implemented across WastewaterSCAN’s national-scale, multi-disease WBE monitoring network. WastwaterSCAN also has led the global WBE research community in investigating issues affecting the performance of WBE methods, in enhancing how WBE data get reported, and in expanding WBE monitoring beyond COVID-19 to about a dozen other diseases. Public health agencies and wastewater treatment operators rely on the WBE data and insights developed, vetted and published by WastewaterSCAN as the scientific foundation for incorporating wastewater monitoring into their routine disease monitoring activities.
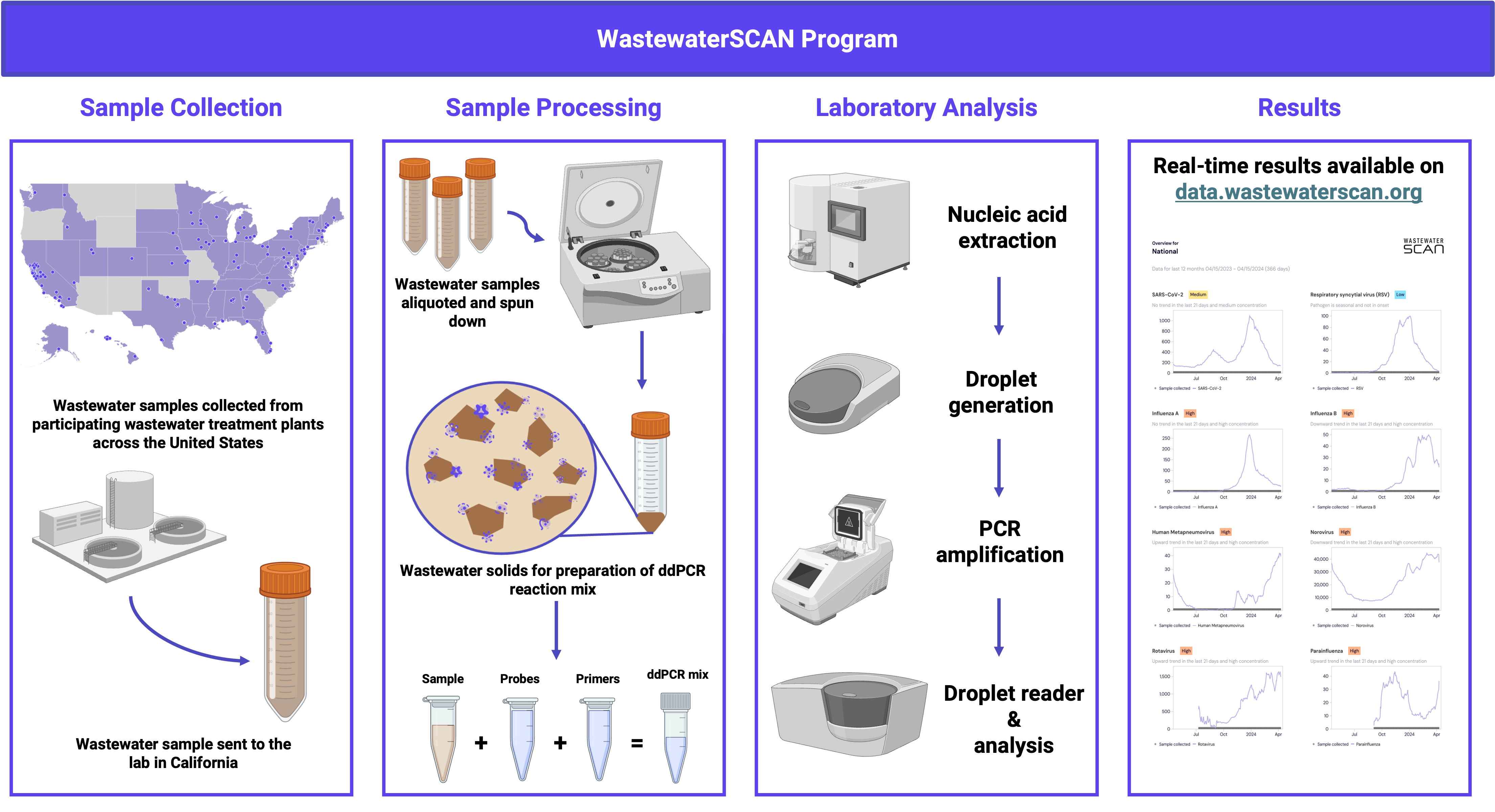
1.1 Leader in developing standardized wastewater processing and analysis protocols
Although the first WBE methods were developed in the early 20th century to monitor polio in wastewater streams, WBE did not gain widespread traction in the U.S. until the COVID-19 pandemic. During the early days of the pandemic, WastewaterSCAN published foundational laboratory protocol papers and studies that brought standardization, consistency and best practices to wastewater monitoring methods – initially, for SARS-CoV-2 monitoring and, subsequently, for a range of other disease targets. This section highlights the pre-analytical processing, nucleic acid extraction, and PCR inhibitor removal techniques and quantification methods that WastewaterSCAN vetted and standardized to guide public health agencies and wastewater treatment plants in extracting reliable, relevant public-health insights from wastewater monitoring. This body of published work remains a preeminent, authoritative source of information for how to conduct routine wastewater monitoring, with the top papers in this section cited more than 300 times.
1.1.1 Published laboratory protocols
To help public health agencies readily access wastewater sampling, processing and analysis protocols, WastewaterSCAN has published its laboratory protocols as standalone, open-source publications. Although not peer-reviewed, the protocols in this section were vetted and standardized via the peer-reviewed studies summarized in the next section (Section 1.1.2).
High Throughput pre-analytical processing of wastewater settled solids for SARS-CoV-2 RNA analyses
- Analysis: Describes the steps for pre-analytical processing of primary settled solids from wastewater treatment plants for downstream nucleic acid purification and quantification.
- Analysis: Describes the steps for purification of nucleic acids from wastewater solids and preparation for downstream quantitative analysis with Reverse Transcriptase droplet digital Polymerase Chain Reaction (RT-ddPCR).
High Throughput SARS-COV-2, PMMOV, and BCoV quantification in settled solids using digital RT-PCR
- Analysis: Describes the steps for quantitative analysis of nucleic acid from SARS-CoV-2 with a triplex Reverse Transcriptase droplet digital Polymerase Chain Reaction (RT-ddPCR) assay targeting the N Gene, S Gene and ORF1a and a duplex assay targeting Bovine Coronavirus Vaccine (BCoV) and Pepper Mild mottle virus (PMMoV) in extracted and purified RNA samples from solid wastewater samples for population level SARS-CoV-2 community surveillance.
- Analysis: Describes the steps for quantitative analysis of nucleic acid from SARS-CoV-2 with a triplex Reverse Transcriptase droplet digital Polymerase Chain Reaction (RT-ddPCR) assay targeting the N Gene, S Gene and mutation assays (one each for HV69-70, E484K/N501Y, del156-157/R158G, del143-145, and LPPA24S) in extracted and purified RNA samples from solid wastewater samples for population level SARS-CoV-2 community surveillance.
1.1.2 Peer-reviewed publications reflecting laboratory protocol development
The wastewater sampling, processing and analysis protocols that WastewaterSCAN developed and published (Section 1.1.1) were tested and vetted during foundational WastewaterSCAN studies that are highlighted in this section.
Publication date: September 2021
- Novel Laboratory Analysis: Developed a rapid and reliable method for ongoing COVID-19 monitoring, that is sensitive for detection of low concentrations of SARS-CoV-2, scalable to generate data quickly, and comparable across laboratories. In this study, SARS-CoV-2 RNA in wastewater solids was monitored daily at eight wastewater treatment plants and results were posted to a website within 24 hours.
- Key Findings: Found a strong association between SARS-CoV-2 RNA in solids and incident case rates within sewersheds and across sewersheds
- Public Health Takeaways: Monitoring efforts for SARS-CoV-2 RNA in wastewater solids can be scaled up to produce high-frequency (daily), rapid (<24 hours), and sensitive (>1/100,000 COVID-19 incidence) results that correlate with COVID-19 incidence in the community.
Publication date: April 2022
- Novel Laboratory Analysis: New assays were developed and tested to detect specific mutations associated with SARS-CoV-2 variants, including Mu, Beta, Gamma, Lambda, Delta, Alpha, and Omicron, in wastewater settled solids. These novel assays can yield results in less than 24 hours and can be quickly adapted to monitor emerging variants in communities.
- Key Findings: Wastewater analysis reflected the rise, fall, and succession of variants over time, allowing exploration of distinct variant patterns.
- Public Health Takeaways: The detection of specific variant mutations in wastewater samples correlated with the regional occurrence of clinical cases, suggesting we can gain valuable insights into variant circulation in our communities.
Publication date: March 2023
- Novel Laboratory Analysis: This study developed and validated novel hydrolysis probe-based RT-PCR assays that target respiratory viral genomes (including respiratory syncytial virus A and RSV B, influenza A and influenza B, human metapneumovirus, human parainfluenza (1–4), seasonal human coronaviruses, and human rhinovirus RNA) and then applied the assays to wastewater solids collected three times per week at a wastewater treatment plant over 17 months during the COVID-19 pandemic.
- Key Findings: Wastewater analysis reflected the rise, fall, and succession of variants over time, allowing exploration of distinct variant patterns.
- Public Health Takeaways: This study fills the knowledge gaps identified in the literature review by providing measurements of concentrations of various respiratory viruses in wastewater solids including human rhinovirus, parainfluenza, metapneumovirus, influenza A and influenza B, RSV A and RSV B, and seasonal coronaviruses, and showing that their concentrations are associated with traditional measures of disease occurrence in the community.
Trends of Enterovirus D68 Concentrations in Wastewater, California, USA, February 2021–April 2023
Publication date: November 2023
- Novel Laboratory Analysis: An enterovirus D68 (EVD68) assay that detects the VP1 gene was developed to test biweekly wastewater solid samples from two California wastewater treatment plants over the course of 26 months, retrospectively.
- Key Findings: Wastewater EVD68 RNA concentration trends in this study matched the trends in the state case data, noting that EVD68 is not a reportable disease for state health departments. Weekly median wastewater EVD68 RNA concentrations positively correlated with weekly case counts.
- Public Health Takeaways: Wastewater surveillance results are specific for EV-D68 and available 24 hours after sample collection, early warning of EV-D68 levels could be available irrespective of clinical testing. Wastewater surveillance for EV-D68 can inform public health action, including when to issue alerts to improve clinical recognition of the potential for severe respiratory illnesses.
Publication date: May 2024
- Novel Laboratory Analysis: Developed and validated a hydrolysis-probe RT-PCR assay for quantification of the H5 hemagglutinin gene. This assay was applied retrospectively to samples from four WWTPs where springtime Influenza A (IAV) increases were identified and one WWTP where they were not.
- Key Findings: The H5 marker was detected at all four WWTPs coinciding with the increases and not detected in the WWTP without an increase. Positive WWTPs are located in states with confirmed outbreaks of highly pathogenic avian influenza, H5N1 clade 2.3.4.4b, in dairy cattle. Concentrations of the H5 gene approached overall influenza A virus gene concentrations, suggesting a large fraction of influenza virus inputs were H5 subtypes.
- Public Health Takeaways: This work shows that wastewater monitoring can provide an early warning for outbreaks likely to produce contributions to the sewershed outside of the expected human-associated inputs, including for animal outbreaks of diseases with zoonotic potential.
Publication date: July 2024
- Analysis: Implemented a novel assay for C. auris detection on a nationwide scale prospectively from September 2023 to March 2024, analyzing a total of 13,842 samples from 190 wastewater treatment plants across 41 U.S. states.
- Key Findings: C. auris was detected in the wastewater solids from 65 sites (34.2%) with 1.45% of all samples having detectable levels of C. auris nucleic acids. Found that C. auris detections are more frequent in states with older populations and in sewersheds with higher numbers of nursing homes and hospitals
- Public Health Takeaways: Wastewater detections are consistent with demographic factors relevant to C. auris epidemiology like age and number of hospitals or nursing homes. This study highlights the capability of wastewater testing to serve as a sentinel surveillance mechanism for emerging pathogens such as C. auris.
1.2 Demonstrating the advantages of monitoring solids instead of liquids
Wastewater is a complex mixture of liquids and solids; the latter can be fecal in origin or represent other solid waste material that goes down drains and toilets. Most wastewater monitoring efforts have chosen to analyze SARS-CoV-2 present in liquid wastewater samples. WastewaterSCAN showed that SARS-CoV-2 and other viruses being monitored in wastewater accumulate in wastewater solids – and since then, WastewaterSCAN’s national WBE monitoring network has measured only the solid fraction of wastewater. Two advantages of monitoring solids instead of liquids are twofold: (1) The RNA from pathogens such as SARS-CoV-2 tends to be found at significantly higher concentrations in wastewater solids than liquids (Section 1.2.1), and (2) the RNA tends to persist for a long time (Section 1.2.2). The papers in this section reflect the key WastewaterSCAN studies that document both of these advantages.
1.2.1. Higher pathogen concentrations in solids
To be useful as a disease monitoring tool, wastewater samples ideally need to concentrate as much nucleic-acids from pathogens as possible. Higher concentrations of pathogens of interest enable monitoring programs to both collect smaller sample sizes and simultaneously maximize the chances of detecting and measuring target pathogens. Initially, WastewaterSCAN investigated if RNA from SARS-CoV-2 is more concentrated in wastewater solids vs. liquids. WastewaterSCAN showed that the nucleic-acids from SARS-CoV-2 and other disease targets preferentially attach, or adsorbs, to wastewater solids in a process known as partitioning. In fact, WastewaterSCAN found that the nucleic-acids are orders of magnitude more concentrated, or enriched, in wastewater solids than liquids on a mass-equivalent basis. These findings are consistent with previous work that has shown that viruses tend to accumulate on solids in both wastewater and in natural environments (Templeton et al. 2008; Rao et al. 1984; Ye et al. 2016; Peccia et al. 2020). The papers in this section highlight WastewaterSCAN’s work to demonstrate that this phenomenon is consistent across SARS-CoV-2, RSV, and rhinovirus nucleic-acids.
Publication date: February 2022
- Analysis: Six laboratories assessed the concentrations of SARS-CoV-2 RNA in the liquid and solid fractions of wastewater from five wastewater treatment plants over two to six months.
- Key Findings: SARS-CoV-2 concentrations measured in solids and liquids were positively correlated and were higher in the solids fractions on a mass equivalent basis by three orders of magnitude.
- Public Health Takeaways: These results utilizing wastewater solids can inform the design of wastewater monitoring programs aimed to better understand the incidence and epidemiology of COVID-19.
Publication date: August 2023
- Analysis: Examined the solid–liquid partitioning behavior of four viruses in wastewater: SARS-CoV-2, respiratory syncytial virus (RSV), rhinovirus (RV), and F+ coliphage/MS2.
- Key Findings: SARS-CoV-2, respiratory syncytial virus, rhinovirus, and MS2 RNA was enriched in the solid fraction (defined as material generally larger than 0.3 μm in hydrodynamic diameter) of wastewater over the liquid fraction in actual wastewater samples. Adsorption coefficients were determined for the viruses in laboratory experiments.
- Public Health Takeaways: This study fills an important knowledge gap on the partitioning of respiratory viruses in wastewater and indicates that they partition preferentially to the wastewater solids. Respiratory virus wastewater surveillance programs could target wastewater solids to improve measurement sensitivity.
Publication date: May 2024
- Analysis: Determined the partition coefficient of dengue, West Nile, Zika, hepatitis A, influenza A, and SARS-CoV-2 viruses in wastewater by conducting a series of batch experiments, where we spiked different concentrations of each virus to wastewater samples from eleven wastewater treatment plants across the United States.
- Key Findings: Viral RNA concentrations were orders of magnitude higher in solids than in the liquid fraction of wastewater samples, for all viruses analyzed in this study.
- Public Health Takeaways: Found that viral markers can be highly enriched in solids, which underscores the importance of considering wastewater solids as a matrix for early detection and monitoring of viral infectious diseases, particularly in communities with low levels of infections.
1.2.2. Long pathogen persistence in solids
To be useful as a disease monitoring tool, the nucleic-acids from pathogens need to persist for as long as possible in wastewater streams. Once these nucleic acids degrade to a point where they are no longer detectable, WBE ceases to be effective as a tool for detecting infectious diseases in wastewater. WastewaterSCAN found that viral nucleic acids can be highly persistent in primary settled solids for several weeks and even months, depending on temperature conditions. Moreover, wastewater solid samples can be stored and then later analyzed via a process known as biobanking. Biobanking has paved the way for retrospective studies and analyses that have provided public health agencies with key insights for understanding emerging viral outbreaks. The papers in this section chronicle WastewaterSCAN’s work to demonstrate the relative persistence of RNA in wastewater liquids vs. solids.
Persistence of endogenous SARS-CoV-2 and pepper mild mottle virus RNA in wastewater settled solids
Publication date: March 2022
- Analysis: Primary settled solids samples were stored for 10 days at three temperatures (4, 22, and 37 °C), which represent typical environmental conditions from cold, temperate, and tropical regions, to assess the effects of temperature and wastewater treatment plant on the decay rate constants and compared the decay rate constants for SARS-CoV-2 RNA in primary settled solids to previously reported decay rate constants in wastewater.
- Key Findings: SARS-CoV-2 and PMMoV genomic RNA is highly stable in wastewater settled solids over 10 days at several environmentally relevant temperatures.
- Public Health Takeaways: Results from this study and previous studies of SARS-CoV-2 RNA in wastewater suggest limited decay of viral nucleic acids during their transit through the sewer network.
Persistence of Human Respiratory Viral RNA in Wastewater Settled Solids
Publication date: March 2024
- Analysis: Investigated the decay kinetics of genomic nucleic-acids of seven human respiratory viruses, including SARS-CoV-2, RSV, human coronavirus (HCoV)-OC43, HCoV-229E, HCoV-NL63, human rhinovirus (HRV), and influenza A virus, as well as pepper mild mottle virus (PMMoV) in wastewater solids.
- Key Findings: Study showed limited decay of viral RNA targets typically measured for respiratory disease WBE. Found that prolonged storage at 4°C for up to 50 days will have a negligible impact on concentrations of the tested viral targets and that decay rates increase with higher temperatures (37°C).
- Public Health Takeaways: The decay rate constants provided in this study for viral RNA in wastewater solids can be used directly in fate and transport models to inform the interpretation of measurements made during wastewater surveillance.
1.3 Leader in investigating factors affecting performance of wastewater methods
Before public health agencies can use WBE data to inform decision-making, they need to know how various factors can affect the performance of wastewater methods. This context helps public health agencies to be aware of any potential limitations or caveats that can affect WBE data and how WBE data are interpreted, including if and how variability in wastewater collection, processing and analysis methods affects results. WastewaterSCAN has emerged as a leader in investigating how different factors associated with wastewater sampling, processing and analysis methods affect data quality. The papers in this section highlight WastewaterSCAN’s efforts to understand how wastewater monitoring data are affected by four factors: storage of wastewater samples, sensitivity of WBE assays, pathogen shedding from human excretions, and sampling frequency.
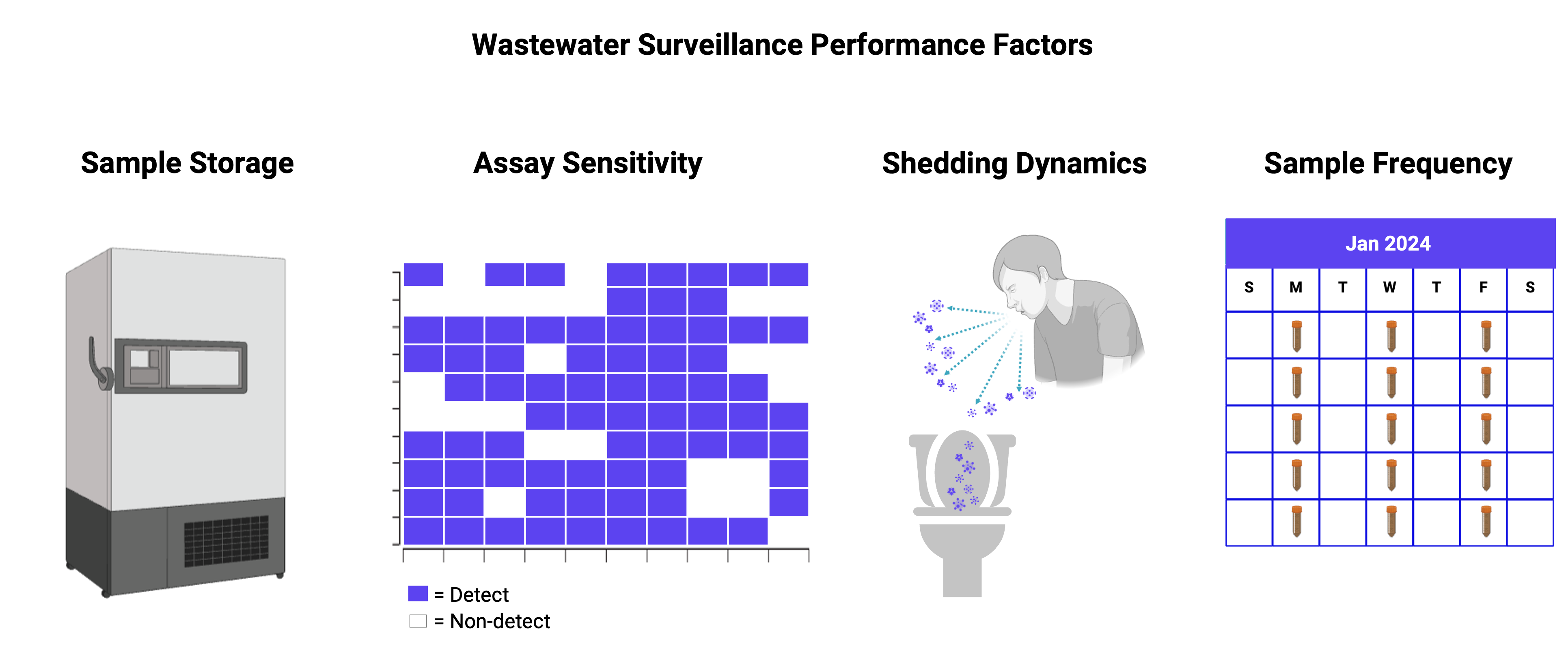
1.3.1 Sample storage
How wastewater samples are stored between the time when they are collected and when they are processed can have a consequential influence on WBE data quality. WastewaterSCAN has shown that degradation of WBE monitoring targets can start as soon as the sample is collected, but that degradation in the solids component of wastewater can be minimal. This finding underscores the advantage of monitoring solids, especially for efforts that involve biobanking solids samples for future analysis. The paper in this section illuminates for public health agencies how sample storage affects the persistence of WBE monitoring targets.
Effect of storage conditions on SARS-CoV-2 RNA quantification in wastewater solids
Publication date: August 2021
- Analysis: This study investigated how storage of wastewater solids at 4 °C, −20 °C, and −80 °C affects measured concentrations of SARS-CoV-2 RNA
- Key Findings: Short term storage (7-8 days) of wastewater solids at 4°C has little effect on SARS-CoV-2 RNA concentrations. However, longer-term storage (35-122 days) at 4°C or freezing at -20°C or –80°C reduces RNA concentrations by an average of 60%. Normalizing SARS-CoV-2 RNA concentrations by the concentration of pepper mild mottle virus (PMMoV) can correct for changes in concentration caused by storage, as storage conditions can affect PMMoV RNA concentrations in a similar manner.
- Public Health Takeaways: Storing wastewater solids may slightly reduce SARS-CoV-2 RNA concentrations, but changes are generally minor, even with extended storage exceeding 100 days. This is especially in contrast to other studies reporting significant reductions in concentrations within liquid wastewater samples under similar storage conditions.
1.3.2 Assay sensitivity
The sensitivity of a WBE assay dictates the lowest levels of a disease target that can be detected using the assay – a level known as the limit of detection. Thus, understanding assay sensitivity is critical to understand when evaluating the overall limitations of WBE as a disease monitoring tool. The paper in this section highlights how to increase the sensitivity of WBE molecular assays that target viral RNA.
Publication date: November 2022
- Analysis: The molecular assay sensitivity of SARS-CoV-2 using digital droplet RT-PCR was examined to understand how it can affect its wastewater-based epidemiology when COVID-19 incidence varies.
- Key Findings: Assays were more sensitive to detecting SARS-CoV-2 RNA at low concentrations (< 104 cp/g) when running 6 or more wells and then more sensitive at high concentrations when running 3 or more wells.
- Public Health Takeaways: Results support an adaptive approach where assay sensitivity is increased by running 6 or more wells during periods of low SARS-CoV-2 gene concentrations, and 3 or more wells during periods of high SARS-CoV-2 gene concentrations.
1.3.3 Shedding of pathogens from human excretions
To track disease occurrence in sewersheds using WBE data, public health agencies need to be able to translate their WBE data into estimates of disease occurrence. One key factor that influences this translation step is how much of a disease target is actually shed into wastewater via human excretions. WastewaterSCAN has played a key role in quantifying shedding rates for multiple disease targets, as well as quantifying the level of uncertainty associated with these estimates. Public health agencies rely on this information to plug into WBE models to estimate disease occurrence. The papers in this section reflect WastewaterSCAN’s efforts to quantify shedding rates for multiple viral pathogens, as well as the uncertainty associated with these estimates – information that can then be directly incorporated into WBE models.
Publication date: October 2021
- Analysis: This study sought to provide standardized methods to preserve stool, and to extract and quantify viral RNA. Stool samples were stored using a variety of storage preservatives and RNA was extracted using 3 different extraction methods to test the amount of detectable RNA preserved.
- Key Findings: Identify that the Zymo DNA/RNA preservative and the QiaAMP extraction kit yield more detectable RNA than the others, using both ddPCR and RT-qPCR.
- Public Health Takeaways: Demonstrated experimental strategies that enable the adoption of the more accessible RT-qPCR assay to enable the accurate detection and relative quantification of viral load in samples. Also recommend reporting quantified viral RNA load in terms of copies per gram of stool. This enables a normalized dataset that will allow researchers to harmonize reported fecal viral loads of SARS-CoV-2 RNA across studies.
Publication date: May 2023
- Analysis: Conducted a systematic review and meta-analysis to characterize the concentrations and presence of influenza A and B, respiratory syncytial virus, metapneumovirus, parainfluenza virus, rhinovirus, and seasonal coronaviruses in stool, urine, mucus, sputum, and saliva. Identified 220 data sets from 50 articles and reported viral concentrations and presence in these excretions
- Key Findings: There is limited data for shedding of respiratory viruses in human excretions, particularly stool and urine samples. The vast majority of excretion data for respiratory viruses is for sputum, mucus, and saliva, which are not the largest theorized contributors to wastewater concentrations.
- Public Health Takeaways: Studies have established that many respiratory viruses are shed in the excretions that comprise wastewater, but further work is needed to generate robust data sets of viral concentrations in these excretions, especially over the course of a respiratory infection.
Publication date: June 2023
- Analysis: Longitudinal shedding of SARS-CoV-2, PMMoV, and crAssphage in stool samples of 48 SARS-CoV-2 positive patients was measured over a minimum of 14 days.
- Key Findings: Of all individuals, 73% shed SARS-CoV-2 in stool, and 51% of all stool samples collected were positive for SARS-CoV-2.Fecal shedding peaked at 2 days from symptom onset.
- Public Health Takeaways: Data presented in this study provide externally valid and longitudinal fecal shedding data for SARS-CoV-2, PMMoV, and crAssphage which can be directly applied to WBE models and ultimately increase the utility of WBE.
1.3.4 Sampling frequency
The gold standard for wastewater monitoring is to sample daily, as this frequency ensures that WBE data can be used to generate statistically significant insights. But daily sampling is not always feasible. Thus, public health agencies find value in understanding the minimum frequencies at which they need to sample to track disease trends accurately. The paper in this section summarizes the guidance that WastewaterSCAN has developed on recommended sampling frequencies for WBE monitoring.
Publication date: April 2023
- Analysis: To identify trends in wastewater monitoring data, this study applied three trend analysis methods (relative strength index (RSI), percent change (PC), Mann-Kendall (MK) trend test) to daily measurements of SARS-CoV-2 RNA in wastewater solids from a wastewater treatment plant to characterize trends.
- Key Findings: Percent change and Mann-Kendall are better indicators of wastewater COVID-19 trends than relative strength index analysis. Downsampling analysis demonstrated a minimum need for 4 samples/week to estimate trends similar to those that would have been measuring using daily data.
- Public Health Takeaways: WBE programs can adopt these trend analysis approaches and sampling frequency recommendations to better inform public health departments how COVID-19 cases are changing, especially as rates of clinical testing continue to decline.
1.4 Leader in bringing standardized best practices to QA/QC and data reporting
As with all scientific data, researchers need to invest time into developing best practices for data QA/QC (quality assurance/quality control) and data reporting. WastewaterSCAN has participated in efforts within the WBE scientific community to develop consensus around standardized best practices for ensuring that WBE monitoring programs around the world are producing high-quality, comparable data sets and making them readily and widely accessible to other researchers. The papers in this section describe WastewaterSCAN’s efforts to bring robust data practices to WBE monitoring.
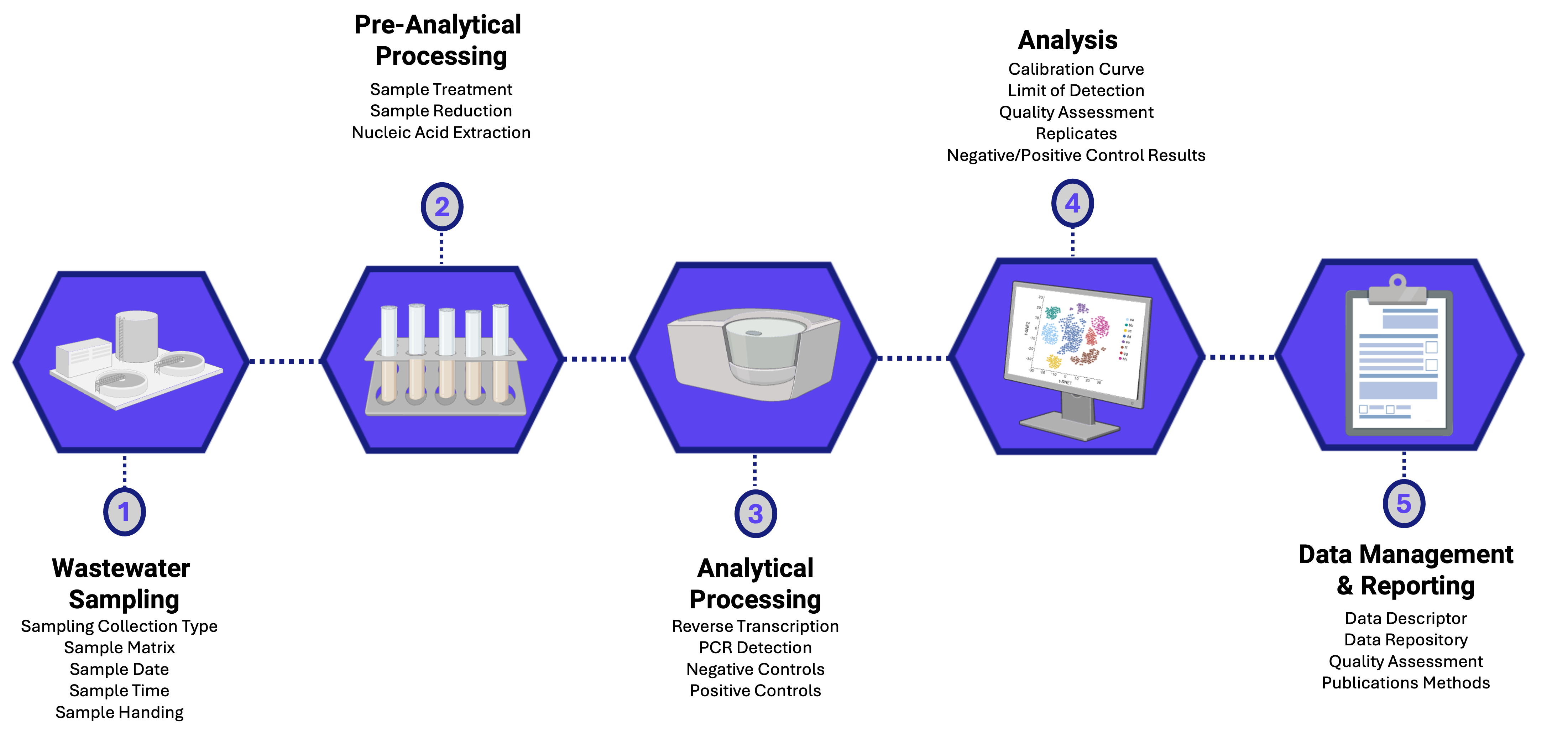
Publication date: July 2021
- Workshop Goal: To establish a recommendation on minimum information to accompany reporting of SARS-CoV-2 occurrence in wastewater for the research community, the United States National Science Foundation (NSF) Research Coordination Network on Wastewater Surveillance for SARS-CoV-2 hosted a workshop in February 2021 with participants from academia, government agencies, private companies, wastewater utilities, public health laboratories, and research institutes.
- Key Findings: Recommendations made include 1) a list with the minimum set of information that should be included with SARS-CoV-2 wastewater measurements and 2) make their data publicly available whenever feasible, ideally through deposition into public repositories, as this can greatly facilitate efficient technology development and method optimization.
- Public Health Takeaways: The framework presented in this publication can serve as a basis for harmonizing data reporting across applications for academic researchers, government agencies, and private companies.
Publication date: July 2021
- Analysis: Describe the application, utility, and interpretation of the suite of controls needed to make high quality qPCR and dPCR measurements of microorganisms in the environment.
- Key Findings: Developed a list of process steps, controls, matrices, and critical reporting categories as an aid for each laboratory to develop their own reporting framework for controls and quality assurance elements.
- Public Health Takeaways: Following the EMMI and MIQE guidelines improves comparability and reproducibility among research studies, better informs crucial engineering and public health decisions, enhances the translation and relevance of research findings, and for our most pressing problems, strengthens the weight of evidence in the direction toward solutions.
Preventing Scientific and Ethical Misuse of Wastewater Surveillance Data
Publication date: August 2021
- Public Health Takeaways: Wastewater surveillance provides an opportunity for wastewater to play a new, dynamic role in the protection of public health in our communities. Additional ethical guidance related to the protection of privacy, balancing individual rights with population interests, and the communication and use of findings is needed.
Publication date: November 2022
- About the datasource: Concentrations of SARS-CoV-2, influenza A and B virus, RSV, mpox virus, norovirus GII, and pepper mild mottle virus nucleic acids were measured at 12 treatment plants in Central California in wastewater solids for up to two years.
- Key Findings: The data generated in the study are valuable for predicting disease trends and hospitalizations in communities and emphasize the potential of wastewater surveillance as a tool for understanding disease epidemiology. Rigorous quality control measures were taken to ensure data accuracy.
- Public Health Takeaways: This publicly available dataset can be used to better understand disease occurrence in communities contributing to the wastewater.
Publication date: October 2024
- About the datasource: Concentrations of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and its variants, influenza A and B viruses, respiratory syncytial virus, human metapneumovirus, enterovirus D68, human parainfluenza types 1, 2, 3, 4a, and 4b in aggregate, norovirus genotype II, rotavirus, Candida auris, hepatitis A virus, human adenovirus, mpox virus, H5 influenza A virus, and pepper mild mottle virus nucleic acids were measured in wastewater solids prospectively at 191 wastewater treatment plants in 40 states across the United States plus Washington DC. Measurements were made two to seven times per week from 1 January 2022 to 30 June 2024, depending on wastewater treatment plant staff availability.
- Key Findings: The data generated in the study are valuable for predicting disease trends and hospitalizations in communities and emphasize the potential of wastewater surveillance as a tool for understanding disease epidemiology. Rigorous quality control measures were taken to ensure data accuracy.
- Public Health Takeaways: This publicly available dataset can be used to better understand disease occurrence in communities contributing to the wastewater.
2. Using WBE to generate public health insights
WastewaterSCAN is leveraging its own WBE monitoring network of 190+ sites across 41 U.S. states, including Washington, D.C., to conduct foundational, proof-of-concept studies demonstrating how public health agencies can generate a range of actionable insights from wastewater data. The studies, which range from national-scale analyses to regional and site-specific studies, have helped build the public health community’s confidence in using WBE methods for routine disease monitoring. Moreover, the studies have demonstrated how WBE can be used as a complement to traditional clinical data indicators, providing a vitally important second line of evidence for tracking disease occurrence in communities. WastewaterSCAN studies also have shown that wastewater data can be used to generate additional types of insights that aren’t possible with clinical data, and to generate these insights earlier and faster than clinical data.
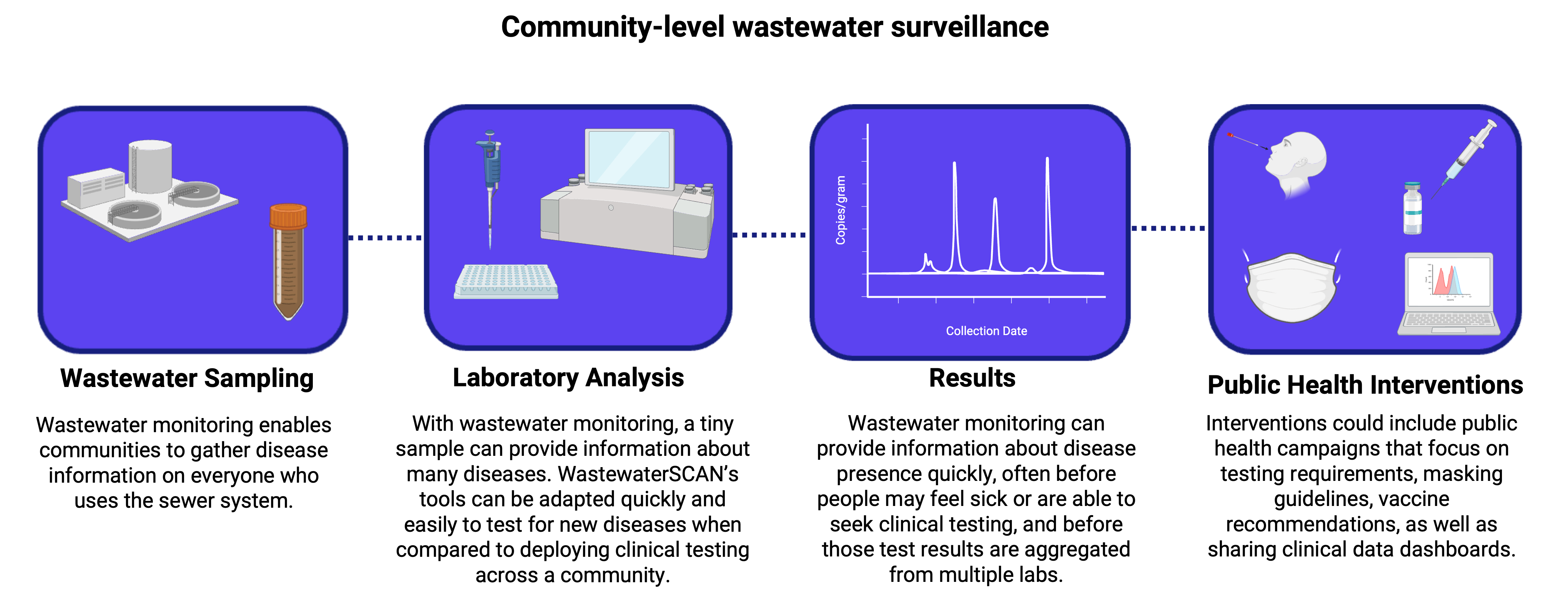
2.1. Correlating WBE data and public health clinical data
Before public health agencies can begin using WBE data as a routine tool for monitoring a disease, they need to know that the wastewater data they are generating reflect disease trends in the underlying population. Scientists investigate this question by conducting correlation studies that help the public health community understand the relationship between insights generated via wastewater data and clinical insights generated via established surveillance methods (e.g., local case counts and test positivity rates). The papers in this section describe the range of correlation studies that WastewaterSCAN has undertaken to give public health agencies confidence about using wastewater as a monitoring and decision-making tool.
2.1.1. Establishing correlations by disease
Different infectious diseases can follow very different trajectories during outbreaks, so scientists must perform correlation analyses for each disease that public health agencies are interested in tracking via wastewater. WastewaterSCAN has demonstrated that clinical case data correlate strongly with wastewater for nearly a dozen disease targets. WastewaterSCAN has conducted much of this work at the sewershed level, which is a geographic area that encompasses all of the public sewer lines that flow to a single end point – in this case, a wastewater treatment plant. WastewaterSCAN also has aggregated data from multiple sewersheds to conduct regional, state and national-scale correlation analyses, further extending the confidence that public health agencies can place in using wastewater data to track disease trends. The papers in this section reflect the correlation analysis work that WastewaterSCAN has completed for respiratory pathogens, including SARS-CoV-2 and its variants, enteric viruses, and a range of other pathogens of concern.
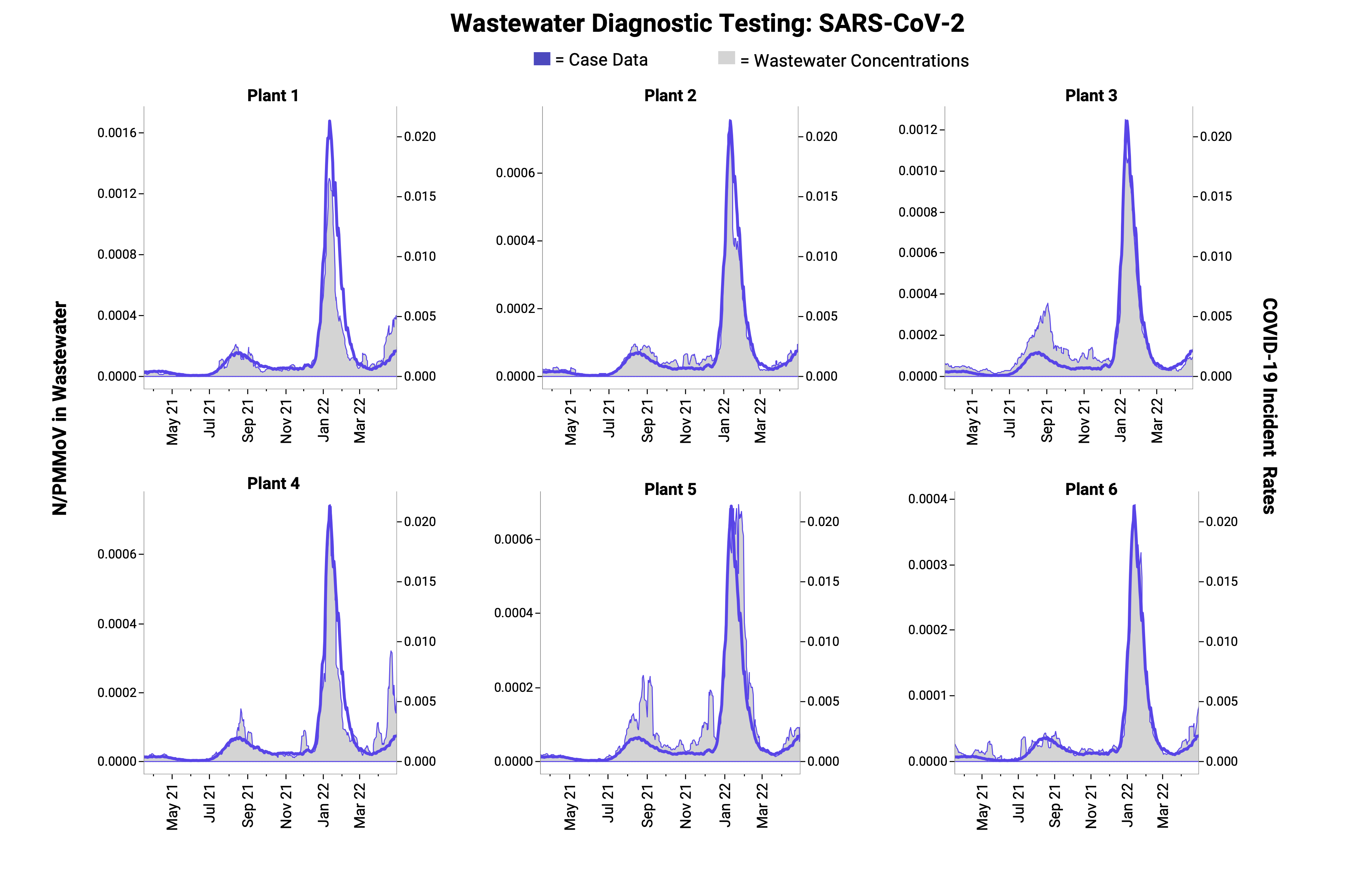
SARS-CoV-2:
Publication date: September 2021
- Analysis: Developed a rapid and reliable method to be useful for ongoing COVID-19 monitoring, that is sensitive for detection of low concentrations of SARS-CoV-2, scalable to generate data quickly, and comparable across laboratories. In this study, SARS-CoV-2 RNA in wastewater solids was monitored daily at eight wastewater treatment plants and results were posted to a website within 24 hours.
- Key Findings: Found a strong association between SARS-CoV-2 RNA in solids and incident case rates within sewersheds and across sewersheds.
- Public Health Takeaways: Monitoring efforts for SARS-CoV-2 RNA in wastewater solids can be scaled up to produce high-frequency (daily), rapid (<24 hours), and sensitive (>1/100,000 COVID-19 incidence) results that correlate with COVID-19 incidence in the community.
Publication date: December 2020
- Analysis: To measure the signal of SARS-CoV-2 in municipal wastewater, there are two key methodological decisions that were compared: 1) The matrix to use: liquid wastewater at the influent of the plant or solids that are concentrated and settled during treatment, and 2) The assay used to quantify the amount of extracted RNA in the sample a: RT-qPCR or digital droplet RT-PCR (ddRT-PCR)
- Key Findings: SARS-CoV-2 RNA was most readily detected when RNA was extracted from wastewater settled solids and quantified with ddRT-PCR. The SARS-CoV-2 RNA measured in wastewater solids correlated positively and significantly with COVID-19 clinical case data from the sewershed over 89 days of measurement.
- Public Health Takeaways: SARS-CoV-2 RNA in wastewater solids can be monitored to assess the COVID-19 burden in communities.
Publication date: June 2023
- Analysis: Examined the divergence of SARS-CoV-2 RNA in wastewater solids compared to COVID-19 clinical case data starting in 2022.
- Key Findings: The divergence could be explained, in part, by a decrease in laboratory test seeking behavior concurrent with an increase in the availability of at-home antigen tests.
- Public Health Takeaways: SARS-CoV-2 RNA concentrations in wastewater could be used to obtain a less biased estimate COVID-19 cases in the community compared to COVID-19 case data, which are affected by fluctuations in test seeking behavior.
Publication date: June 2024
- Analysis: Utilized wastewater SARS-CoV-2 RNA monitoring data, COVID-19 case surveillance data, and dates associated with changes to campus COVID-19 public health policies between 29 July 2021 to 9 August 2023 to examine public health policy impact on the spread of SARS-CoV-2 on a university campus.
- Key Findings: Did not observe changes in SARS-CoV-2 RNA wastewater concentrations associated with most policy changes. Policy changes associated with a significant change in campus wastewater SARS-CoV-2 RNA concentrations included changes to face covering recommendations, indoor gathering bans, and routine surveillance testing requirements and availability.
- Public Health Takeaways: Longitudinal wastewater monitoring of viruses at a small geographic scale may be used for causal inference when randomized controlled experiments are not possible to conduct.
SARS-CoV-2 variants:
Publication date: April 2022
- Analysis: New assays were developed and tested to detect specific mutations associated with SARS-CoV-2 variants, including Mu, Beta, Gamma, Lambda, Delta, Alpha, and Omicron, in wastewater settled solids. These novel assays can yield results in less than 24 hours and can be quickly adapted to monitor emerging variants in communities.
- Key Findings: Wastewater analysis reflected the rise, fall, and succession of variants over time, allowing exploration of distinct variant patterns.
- Public Health Takeaways: The detection of specific variant mutations in wastewater samples correlated with the regional occurrence of clinical cases, suggesting we can gain valuable insights into variant circulation in our communities.
Publication date: Jan 2021
- Analysis: This paper describes the initial detections in wastewater during November 21–December 16, 2021, and the interpretative framework for these types of data.
- Key Findings: Wastewater data played a crucial role in providing strong early evidence of Omicron’s presence and distribution, and can be used as a complement to clinical testing for early detection of variants and public health decision-making.
- Public Health Takeaways: There are some limitations with current wastewater surveillance methods that limit the detection of variant identification, emphasizing the importance of interpreting data carefully.
Publication date: May 2022
- Analysis: Developed and tested two new assays, one that targets characteristic mutations in the BA.2 sublineage of Omicron and another that targets a set of mutations present in both BA.1 and BA.2. Then applied the assays to wastewater samples collected daily at eight POTW in the greater Bay Area, California, United States.
- Key Findings: Wastewater samples documented regional replacement of BA.1 with BA.2 in agreement with, and ahead of, clinical sequencing data (which typically have a 1-2 week delay).
- Public Health Takeaways: Wastewater measurements obtained using these assays are available within 24 h of receiving samples at the laboratory providing information ahead of clinical sequencing data. Real-time estimates of variant data from wastewater can allow clinicians and regulators to identify the most effective treatment for a particular community.
Publication date: May 2022
- Analysis: Developed targeted digital reverse transcription PCR mutation assays to retrospectively and prospectively monitor wastewater settled solids for the presence and abundance of mutations present in the Alpha (B.1.1.7) and Delta (B.1.617.2) variants of concern. Compared these totals against estimates of Alpha and Delta abundance in two California sewersheds by using COVID-19 case isolate sequencing data available to the California Department of Public Health (CDPH).
- Key Findings: Results show that the HV69-70 and Del156-157/R158G mutation assays as used for wastewater settled solids were sensitive and specific. By using these PCR mutation assays, we found strong correlation between wastewater estimates and case isolate sequencing–derived estimates of circulating Alpha and Delta in 2 large metropolitan communities in California, USA.
- Public Health Takeaways: Mutations were detected in wastewater samples collected 1–3 weeks earlier than when Alpha and Delta variant estimates generated by case-isolate sequencing were available and reliable. Wastewater variant monitoring can overcome biases and delays seen with case isolate sequencing.
Influenza:
Wastewater-based detection of two influenza outbreak
Publication date: July 2023
- Analysis: Influenza (IAV) RNA concentrations in wastewater reflected outbreak patterns and magnitude at two university campuses in different parts of the United States and different times of year
- Key Findings: IAV RNA concentrations were strongly associated with reported IAV incidence rates (Kendall’s τ values of 0.58 and 0.67 for the University of Michigan and Stanford University, respectively).
- Public Health Takeaways: Wastewater surveillance can effectively detect IAV outbreaks and is a valuable supplement to traditional forms of IAV surveillance.
Publication date: November 2023
- Analysis: Influenza A (IAV) RNA was measured in wastewater settled solids at 163 wastewater treatment plants across 33 states to characterize the 2022-2023 influenza season (season onset, offset, duration, peak, and intensity) to compare with those determined using laboratory-confirmed influenza hospitalization rates and outpatient visits for influenza-like illness (ILI).
- Key Findings: IAV RNA in wastewater provided early warning of onset, compared to the ILI estimate, when the baseline was set at twice the limit of IAV RNA detection in wastewater. IAV RNA in wastewater correlated strongly with influenza hospitalization rates.
- Public Health Takeaways: Wastewater settled solids data are an IAV-specific indicator that can be used to augment clinical surveillance for seasonal influenza epidemic timing and intensity.
RSV:
Publication date: January 2022
- Analysis: Designed and tested a custom digital droplet (dd-)RT-PCR assay that targets the N gene of RSV A and RSV B in wastewater solids at two publicly owned treatment works (POTWs) in a highly urbanized region of the San Francisco Bay Area, California, United States.
- Key Findings: RSV genomic RNA can be detected in wastewater solids from wastewater treatment plants using a novel, sensitive and specific RT-PCR assay. The concentrations of RSV RNA in wastewater solids at two wastewater treatment plants correlates strongly with positivity rates from clinical specimens from the state.
- Public Health Takeaways: Environmental monitoring of wastewater solids can provide information on RSV occurrence in the community contributing to wastewater.
Publication date: February 2024
- Analysis: Conducted a retrospective observational analysis using data on RSV RNA concentrations in wastewater collected as part of this study along with publicly available clinical and laboratory data on RSV test positivity and hospital admissions.
- Key Findings: A total of 22,809 samples were collected from 176 WastewaterSCAN sites (data from 1 January 1, 2022 to 31 July 31, 2023 were used). RSV RNA concentrations aggregated at state and national levels correlated with infection positivity and hospitalization rates.
- Public Health Takeaways: Wastewater concentrations are less subject to biases in testing capacity and test-seeking behavior.
EV-D68:
Trends of Enterovirus D68 Concentrations in Wastewater, California, USA, February 2021–April 2023
Publication date: November 2023
- Analysis: An enterovirus D68 (EVD68) assay that detects the VP1 gene was developed to test biweekly wastewater solid samples from two California wastewater treatment plants over the course of 26 months, retrospectively.
- Key Findings: Wastewater EVD68 RNA concentration trends in this study matched the trends in the state case data, noting that EVD68 is not a reportable disease for state health departments. Weekly median wastewater EVD68 RNA concentrations positively correlated with weekly case counts.
- Public Health Takeaways: Wastewater surveillance results are specific for EV-D68 and available 24 hours after sample collection, early warning of EV-D68 levels could be available irrespective of clinical testing. Wastewater surveillance for EV-D68 can inform public health action, including when to issue alerts to improve clinical recognition of the potential for severe respiratory illnesses.
Multiple Respiratory Diseases:
Publication date: July 2023
- Analysis: This study measured concentrations of genomic nucleic acids of influenza A (IAV), RSV, and SARS-CoV-2, as well as human metapneumovirus (HMPV), in wastewater solids daily in eight publicly owned treatment works (POTWs) in the Greater San Francisco Bay Area, California, USA during winter 2022–2023. Also, designed novel hydrolysis probe RT-PCR assays for different IAV subtype markers to identify the dominant circulation IAV subtype (H3N2).
- Key Findings: Identified a “tripledemic” when wastewater concentrations of IAV, RSV and SARS-CoV-2 collected from daily wastewater measurements at eight public wastewater treatment plants (POTW) in the San Francisco Bay Area, CA reflected when cases of IAV, RSV and SARS-CoV-2 increased dramatically. HMPV in wastewater samples differed widely in timing of onset and peak across the eight POTW suggesting localized dynamics in the wastewater events that are not reflected in the state-aggregated clinical data.
- Public Health Takeaways: Wastewater can be used to help identify the onset and offset of respiratory virus transmission events as well as the peak of such events. It can also be used to identify influenza subtypes.
Publication date: March 2023
- Analysis: This study developed and validated novel hydrolysis probe-based RT-PCR assays that target respiratory viral genomes (including respiratory syncytial virus (RSV) A and RSV B, human metapneumovirus, human parainfluenza (1–4), seasonal human coronaviruses, and human rhinovirus RNA) and then applied the assays as well as existing assays for influenza A and B to wastewater solids collected three times per week at a wastewater treatment plant over 17 months during the COVID-19 pandemic.
- Key Findings: Wastewater analysis reflected the rise, fall, and succession of viruses over time, and matched state-level data on infection positivity.
- Public Health Takeaways: This study provides measurements of concentrations of various respiratory viruses in wastewater solids including human rhinovirus, parainfluenza, metapneumovirus, influenza A and influenza B, RSV A and RSV B, and seasonal coronaviruses, and shows that their concentrations are associated with traditional measures of disease occurrence in the community.
Enteric viruses:
Publication date: February 2024
- Analysis: Nucleic-acids of adenovirus group F, norovirus G1 and GII, rotavirus, and enterovirus were measured for over 2 years in wastewater solids at two plants in the Bay Area of CA using multiplexed assays in digital PCR to compare viral nucleic acid concentrations to positivity rates for viral infections from clinical specimens submitted to a local clinical laboratory to assess concordance between the data sets
- Key Findings: Concentrations for these pathogens were enriched in the solids over raw influent by 1,000 times. Concentrations of adenoviruses, noroviruses and rotavirus correlated significantly with case positivity rates.
- Public Health Takeaways: Provides evidence for the use of wastewater solids for the sensitive detection of enteric virus targets in WBE programs aimed to better understand the spread of enteric disease at a localized, community level without limitations associated with testing many individuals.
Norovirus:
Wastewater Surveillance for Norovirus, California, USA
Publication date: November 2024
- Analysis: Collected Norovirus wastewater data during December 17, 2022–December 17, 2023, from 76 California wastewater utilities, including sites in all 5 California public health officer region.
- Key Findings: Observed positive, statistically significant (p<0.001), moderate-to-strong correlations between California regional and statewide wastewater aggregates and US national and western regional NREVSS test positivity.
- Public Health Takeaways: Wastewater norovirus GII data from California during 2022–2023 correlated well with existing public health surveillance data. The wastewater data provided otherwise unavailable situational awareness, enabled timely identification of distinct California regional norovirus trends, and led to direct public health action, including guiding local outbreak investigations.
Mpox:
Use of Wastewater for Mpox Outbreak Surveillance in California
Publication date: February 2023
- Analysis: Due to an existing routine wastewater monitoring program in California, PCR assay testing of MPOX DNA concentrations in daily settled wastewater solid samples was able to be implemented less than a month after the first confirmed Mpox case in the United States.
- Key Findings: A strong relationship between MPOX DNA concentrations in wastewater solids and the incidence of MPOX infections suggests that wastewater surveillance can be used as a complementary tool by public health agencies and clinicians.
- Public Health Takeaways: Adapting routine wastewater-surveillance infrastructure to monitor for a nonenteric, nonrespiratory virus such as MPOX shows promise for the future use of this method (link to paper appendix with lab methodology) as an adjunct public health tool.
Dengue:
Wastewater Detection of Emerging Arbovirus Infections: Case Study of Dengue in the United States
Publication date: December 2023
- Analysis: Tested wastewater solids for RNA from the 4 dengue virus serotypes at three WWTP in the Miami, Florida area.
- Key Findings: Consistently detected dengue virus 3 RNA at all three WWTP and did not detect the other three Dengue (1, 2, and 4) serotypes.
- Public Health Takeaways: Wastewater detection of dengue virus RNA is possible with as few as 4 laboratory confirmed dengue cases per 1 million people based on publicly available infection data.
Hepatitis A:
Publication date: October 2024
- Analysis: Measured concentrations of Hepatovirus A (HAV) RNA, in samples from 191 wastewater treatment plants spanning 40 US states and the District of Columbia from September 2023 to June 2024 and compared the measurements with traditional measures of disease occurrence.
- Key Findings: Showed that HAV RNA wastewater concentrations are associated with disease occurrence in the population at the national level, within some states, and at the county level in Maine. This is despite the fact that the incident case data may be biased to cases of severe disease and may not include undiagnosed and subclinical cases. Also found that wastewater detection of HAV RNA is associated with socio-economic indicators of vulnerability. This suggests that data obtained via wastewater monitoring may provide insights into the health of these vulnerable populations.
- Public Health Takeaways: Results highlight the potential for wastewater-based epidemiology to be a complementary tool to traditional surveillance for monitoring and controlling HAV transmission.
2.1.2. Establishing correlations at the building, school, and campus level
While most of the correlation analyses between wastewater and clinical data are done at the sewershed level, there also can be public-health value in far more localized analyses, including individual buildings, schools and campuses. WastewaterSCAN has conducted these hyperlocal analyses for both K-12 schools and college campuses. The papers in this section demonstrate how monitoring wastewater at smaller scales than those served by wastewater treatment plants may help in understanding how diseases are spreading and in assisting with localized public health responses.
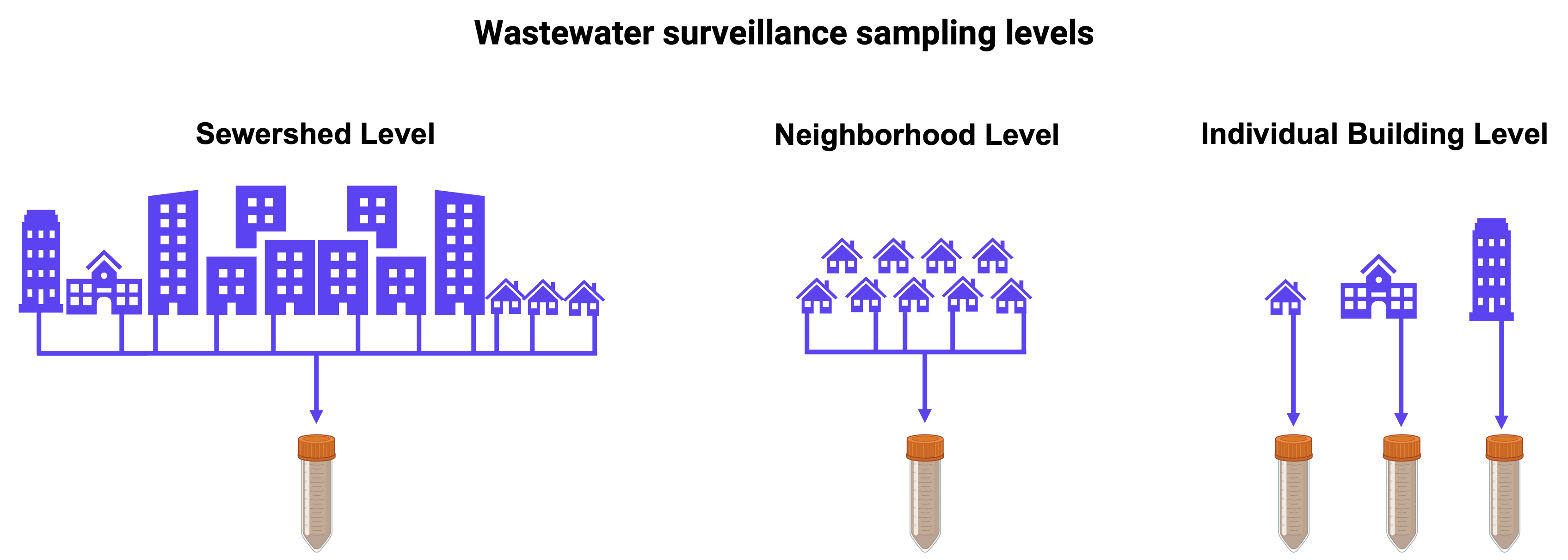
Publication date: May 2022
- Analysis: Sampled wastewater from three nested locations with different sized populations within the same sewer network at a university campus (occurred at the campus level, building cluster level and the individual building level) and quantified SARS-CoV-2 RNA using reverse transcriptase droplet digital PCR.
- Key Findings: SARS-CoV-2 RNA detected at the campus, building cluster and the individual building level and were positively associated with student COVID-19 case data when normalized by PMMoV. Detection of the N antigen using a commercial antigen test at the sewershed level was not associated with laboratory-confirmed COVID-19 cases of SARS-CoV-RNA concentrations in wastewater.
- Public Health Takeaways: Building cluster and building level testing may be primarily useful for investigating the sources of outbreaks and for buildings that have a consistent and known population (e.g., a dormitory or school).
Publication date: October 2022
- Analysis: Campus-level wastewater data was analyzed to determine which SARS-CoV-2 variants were circulating at the university.
- Key Findings: Wastewater samples can be used to infer the circulating variant rather than clinical samples because a large number of students in the cohort never underwent polymerase chain reaction testing, limiting the availability of samples for sequencing.
- Public Health Takeaways: The use of rapid antigen testing to aid in the decision to end isolation may be needed to prevent individuals with infection from leaving isolation prematurely.
Wastewater monitoring of SARS-CoV-2 RNA at K-12 schools: Comparison to pooled clinical testing data
Publication date: March 2023
- Analysis: Clinical data from two California K-12 schools that performed pooled SARS-CoV-2 testing 2-3 times a week had a comparable detection frequency to both the solid and liquid wastewater samples collected within two days of specimen collection.
- Key Findings: Most wastewater samples were positive for SARS-CoV-2 RNA when clinical testing was positive (75% for solid samples and 100% for liquid samples).
- Public Health Takeaways: In similar settings when monitoring SARS-CoV-2 in a sewershed serving a small population with low concentrations in the wastewater, it is recommended to use 1) a very frequent composite sampling scheme at a convenient access point, and 2) multiple PCR targets.
2.2. Demonstrating how to monitor diseases using WBE data only
Once scientists have demonstrated a consistently strong correlation between WBE data and clinical case data for a disease, the public health community can do more than just use WBE monitoring as a second line of evidence complementing traditional clinical disease indicators. Public health agencies also have the option to track diseases using wastewater data only – a potentially valuable option for communities where it is infeasible or time- and cost-prohibitive to collect comprehensive clinical data for one or more disease targets. The papers in this section establish a scientific foundation for how to estimate infectious disease transmissions in real time using wastewater data only.
Publication date: April 2021
- Analysis: Determine how SARS-CoV-2 RNA measurements at different POTW can be used to compare incidence rates of COVID-19 in their sewersheds by developing a mass balance model that links SARS-CoV-2 RNA concentrations in POTW settled solids to the number of individuals shedding SARS-CoV-2 RNA in stool in the sewershed.
- Key Findings: SARS-CoV-2 N1 and N2 concentrations in solids normalized by concentrations of PMMoV RNA in solids can be used to compare incidence of laboratory confirmed new COVID-19 cases across POTWs.
- Public Health Takeaways: Changes in SARS-CoV-2 wastewater concentrations are associated with proportional increases and decreases in cases across different treatment plants in the United States. This means wastewater monitoring concentrations can be compared across treatment plants all over the country, and are an effective predictor of case rates through a broad range of climates and demographics.
Publication date: May 2022
- Analysis: Developed distributed lag models to estimate incidence rate of SARS-CoV-2 based on wastewater concentrations were created for sampling frequencies of once every 2, 3, 4 and 7 days.
- Key Findings: Distributed lag models had strong out-of-sample predictive power for COVID-19 incidence rates using wastewater solids surveillance of SARS-CoV-2 N genes (with lags of 0–3 days) for California sewersheds. The models captured the rise and fall of cases during the introduction of the Delta variant using a model fit to data during a time period with a mixture of variants represented.
- Public Health Takeaways: Incidence rates can be estimated with wastewater sampling as little as every 4 days while retaining limited error rates. However, reduced sampling frequency may not serve other important wastewater surveillance use cases.
Wastewater-based estimation of the effective reproductive number of SARS-CoV-2
Publication date: May 2022
- Analysis: Measured SARS-CoV-2 RNA in sewage sludge or wastewater from two distinct monitoring programs (Zurich, Switzerland, and San Jose, California, USA) to estimate the effective reproductive number (Re - a critical indicator to monitor disease dynamics, inform regional and national policies, and estimate the effectiveness of intervention).
- Key Findings: Reproductive number, the rate at which a viral infection spreads from person to person, can be estimated using wastewater RNA concentrations of SARS-CoV-2. These wastewater based estimates of reproductive number for SARS-CoV-2 match clinical case based estimates.
- Public Health Takeaways: Areas without strong clinical testing programs will be able to use wastewater based estimates of reproductive number to monitor the epidemiological situation in the underlying population with as few as 3 weekly wastewater measurements.
Publication date: July 2022
- Analysis: Developed three mechanistic models for estimating SARS-CoV-2 community infection levels from wastewater measurements that are agnostic to location. Each model uses a distinct approach to account for the human fecal content of wastewater, including endogenous microbial markers, wastewater flow measurements, and population-level estimates of per-capita wastewater generation.
- Key Findings: The resulting infection estimates are location agnostic and match the best estimates of infection based on clinical case rates.
- Public Health Takeaways: Wastewater-based mechanistic models are useful for normalization of wastewater measurements and for understanding wastewater-based surveillance data for public health decision-making but are limited by lack of robust SARS-CoV-2 fecal shedding data.
2.3 Demonstrating WBE’s potential to offer insights that traditional clinical data don’t
While WBE has the potential to serve as a complementary or confirmatory line of evidence for disease monitoring, it also has the potential to provide public health agencies with different types of insights than they can get from traditional clinical data sources. For most diseases, for example, clinical testing is limited or nonexistent. Similarly, clinical testing can be extraordinarily time- and labor-intensive. By contrast, WBE has the potential to readily scale, to capture disease information for a much higher portion of a sewershed’s population, and produce insights more rapidly and cost-effectively than clinical data. The papers in this section summarize WastewaterSCAN’s efforts to demonstrate for the public health community the insights that WBE monitoring can offer that traditional clinical data do not.
Publication date: July 2023
- Analysis: This study measured concentrations of genomic nucleic acids of influenza A (IAV), RSV, and SARS-CoV-2, as well as human metapneumovirus (HMPV), in wastewater solids daily in eight publicly owned treatment works (POTWs) in the Greater San Francisco Bay Area, California, USA during winter 2022–2023. Also, designed novel hydrolysis probe RT-PCR assays for different IAV subtype markers to identify the dominant circulation IAV subtype (H3N2).
- Key Findings: Identified a “tripledemic” when wastewater concentrations of IAV, RSV and SARS-CoV-2 collected from daily wastewater measurements at eight public wastewater treatment plants (POTW) in the San Francisco Bay Area, CA reflected when cases of IAV, RSV and SARS-CoV-2 increased dramatically. HMPV in wastewater samples differed widely in timing of onset and peak across the eight POTW suggesting localized dynamics in the wastewater events that are not reflected in the state-aggregated clinical data.
- Public Health Takeaways: Wastewater can be used to help identify the onset and offset of respiratory virus transmission events as well as the peak of such events.
Publication date: February 2023
- Analysis: HuNoV GII RNA was found to preferentially adsorb to wastewater solids where it is present at 1,000 times the concentration in influent.
- Key Findings: HuNoV GII RNA concentrations in wastewater solids were positively associated with clinical specimen positivity rates both at a single WWTP, and aggregated across WWTPs spanning the entire United States
- Public Health Takeaways: Wastewater measurements were available as quickly as 24 h after a sample is received at a laboratory, so this work suggests that wastewater testing may provide real-time information on changes in disease occurrence for use by public health officials, clinicians, and the public by arriving in databases before data on clinical infections. As there is limited data on disease occurrence for illnesses caused by HuNov, wastewater monitoring can provide information about the community spread of HuNoV.
Publication date: June 2023
- Analysis: Examined the divergence of SARS-CoV-2 RNA in wastewater solids compared to COVID-19 clinical case data starting in 2022.
- Key Findings: The divergence could be explained, in part, by a decrease in laboratory test seeking behavior concurrent with an increase in the availability of at-home antigen tests.
- Public Health Takeaways: SARS-CoV-2 RNA concentrations in wastewater could be used to obtain a less biased estimate COVID-19 cases in the community compared to COVID-19 case data, which are affected by fluctuations in test seeking behavior.
Publication date: Jan 2021
- Analysis: This paper describes the initial detections of Omicron in wastewater during November 21–December 16, 2021, and the interpretative framework for these types of data.
- Key Findings: Wastewater data played a crucial role in providing strong early evidence of Omicron’s presence and distribution, and can be used as a complement to clinical testing for early detection of variants and public health decision-making.
- Public Health Takeaways: There are some limitations with current wastewater surveillance methods that limit the detection of variant identification, emphasizing the importance of interpreting data carefully.
Publication date: May 2022
- Analysis: Developed and tested two new assays, one that targets characteristic mutations in the BA.2 sublineage of Omicron and another that targets a set of mutations present in both BA.1 and BA.2. Then applied the assays to wastewater samples collected daily at eight POTW in the greater Bay Area, California, United States.
- Key Findings: Wastewater samples documented regional replacement of BA.1 with BA.2 in agreement with, and ahead of, clinical sequencing data (which typically have a 1-2 week delay).
- Public Health Takeaways: Wastewater measurements obtained using these assays are available within 24 h of receiving samples at the laboratory providing information ahead of clinical sequencing data. Real-time estimates of variant data from wastewater can allow clinicians and regulators to identify the most effective treatment for a particular community.
Publication date: May 2022
- Analysis: Developed targeted digital reverse transcription PCR mutation assays to retrospectively and prospectively monitor wastewater settled solids for the presence and abundance of mutations present in the Alpha (B.1.1.7) and Delta (B.1.617.2) variants of concern. Compared these totals against estimates of Alpha and Delta abundance in two California sewersheds by using COVID-19 case isolate sequencing data available to the California Department of Public Health (CDPH).
- Key Findings: Results show that the HV69-70 and Del156-157/R158G mutation assays as used for wastewater settled solids were sensitive and specific. By using these PCR mutation assays, we found strong correlation between wastewater estimates and case isolate sequencing–derived estimates of circulating Alpha and Delta in 2 large metropolitan communities in California, USA.
- Public Health Takeaways: Mutations were detected in wastewater samples collected 1–3 weeks earlier than when Alpha and Delta variant estimates generated by case-isolate sequencing were available and reliable. Wastewater variant monitoring can overcome biases and delays seen with case isolate sequencing.
2.4 Supporting and guiding WBE programs worldwide
WastewaterSCAN has played a leading role in helping public health agencies design and implement robust WBE monitoring programs. Not only is WastewaterSCAN’s technical expertise and experience routinely called upon as a scientific adviser and authority figure, but WastewaterSCAN also has demonstrated through its own national-scale WBE monitoring program how to design and operate large-scale monitoring infrastructure. As of 2024, the WastewaterSCAN monitoring network encompassed 190+ sites across 41 U.S. states, including Washington, D.C. All participants are using standardized, best-practices methods for sample collection, processing and analysis, and all monitoring data are publicly published to https://data.wastewaterscan.org as soon as they become available. Through this ongoing monitoring network, WastewaterSCAN has gained the experience and perspective necessary to help guide the evolution and expansion of WBE monitoring around the world. The WastewaterSCAN monitoring network has helped demonstrate the value and relevance of WBE monitoring in communities across the U.S., connected and fostered a sense of community among public health agencies and wastewater treatment plants, and encouraged the free exchange of data, best practices and WBE insights. The papers in this section are peer-reviewed opinion pieces about the value and relevance of WBE monitoring; they reflect WastewaterSCAN’s commitment to encouraging academic researchers and the global public health community to think in broad, visionary terms about the future of WBE monitoring.
Publication date: March 2020
- Public Health Takeaways: Environmental science and engineering researchers should 1) take a broader, long-term, and more quantitative approach to understanding viruses that are spread through the environment, and 2) aim to understand and communicate with colleagues in medicine and public health the specific characteristics that drive transport and inactivation of enveloped viruses in solutions, on surfaces, and in the air.
Wastewater Monitoring Can Anchor Global Disease Surveillance Systems
Publication date: June 2023
- Analysis: Surveyed wastewater surveillance programs in 43 countries to characterize the landscape of global wastewater monitoring.
- Key Findings: Suggest that three core attributes are needed to realize the potential of wastewater surveillance: 1) global and country-level leadership, 2) financial investment, and 3) unifying but flexible implementation frameworks.
- Public Health Takeaways: Many individual wastewater initiatives that now exist can coalesce into an integrated, sustainable network for disease surveillance that minimizes the risk of overlooking or underestimating future emerging global health threats.
Publication date: December 2022
- Public Health Takeaways: There are three interconnected activities that are essential for the support of effective wastewater surveillance systems: 1) pioneering new capabilities, 2) sustaining surveillance efforts, and 3) integrating surveillance data for effective public health response (see figure here). Academic researchers can play an important role in contributing to each of these activities.
Wastewater surveillance for rapid identification of infectious diseases in prisons
Publication date: June 2022
- Public Health Takeaways: Wastewater-based surveillance could enable public health authorities to develop targeted prevention and education programs for hygiene measures and prevention of drug use within prisons.